R Basics
Introduction to Data Science (BIOL7800)
Daijiang Li
LSU
2023/09/12
1 / 25
Data types
The first step in any data analysis is to choose the structure and to create a dataset to hold the data
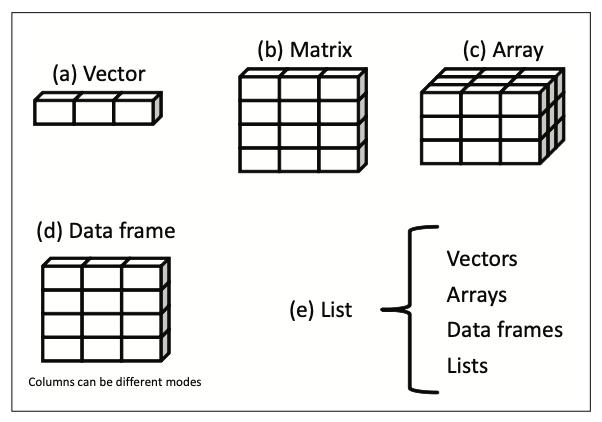
R has a wide variety of structures for holding data, including scalars, vectors, arrays, data frames, and lists.
2 / 25
Data structures
| Dimensions | Homogeneous | Heterogeneous |
|---|---|---|
| 1d | Vector (atomic) | List (generic) |
| 2d | Matrix | Data frame |
| nd | Array | NA |
Almost all other objects are build upon these foundations.
str() to understand data structure
3 / 25
Vector
Vector types: logical, double, integer1 , character, complex (imaginary numbers), and raw (bytes)
Go-to function for making vectors: c()
(a <- c(1:3)) # equal to: a <- c(1:3); a## [1] 1 2 3(b <- c(4:6))## [1] 4 5 6(C <- c(a, b)) # don't name it as c!## [1] 1 2 3 4 5 6[1] double and integer are both numeric
4 / 25
Vector
Vectors have three common properties:
- Type (what it is),
typeof() - Length (how many elements),
length() - Attributes (additional arbitrary metadata)
attributes()
typeof(a)## [1] "integer"length(a)## [1] 3attributes(a)## NULL5 / 25
Vector
(v_dbl = c(1, 3.1))## [1] 1.0 3.1(v_int = c(0L:3L)) # colon operator## [1] 0 1 2 3(v_log = c(TRUE, FALSE)) # T, F## [1] TRUE FALSE(v_chr = c("a", "word"))## [1] "a" "word"6 / 25
Vector
(v_dbl = c(1, 3.1))## [1] 1.0 3.1(v_int = c(0L:3L)) # colon operator## [1] 0 1 2 3(v_log = c(TRUE, FALSE)) # T, F## [1] TRUE FALSE(v_chr = c("a", "word"))## [1] "a" "word"typeof(v_dbl)## [1] "double"is.double(v_dbl)## [1] TRUEis.numeric(v_int)## [1] TRUEis.integer(v_int)## [1] TRUEis.atomic(v_log)## [1] TRUE6 / 25
Coercion
Vector only allow one type of elements; so when mix different types of elements, they will be coerced to the most flexible type (least to most flexible: logical, integer, double, character)
c(v_log, v_int)## [1] 1 0 0 1 2 3c(v_log, v_chr)## [1] "TRUE" "FALSE" "a" "word"c(v_dbl, v_int)## [1] 1.0 3.1 0.0 1.0 2.0 3.0c(v_dbl, v_chr)## [1] "1" "3.1" "a" "word"typeof(c(v_log, v_int))## [1] "integer"typeof(c(v_log, v_chr))## [1] "character"typeof(c(v_dbl, v_int))## [1] "double"typeof(c(v_dbl, v_chr))## [1] "character"7 / 25
Coercion and math functions
Coercion often happens automatically
v_log2 = c(TRUE, FALSE, TRUE, TRUE, FALSE)sum(v_log2)## [1] 3mean(v_log2)## [1] 0.68 / 25
How do you get the number of positive values in the vector below using the coercion example in the previous slide?
v_norm = rnorm(n = 1000, mean = 0, sd = 2)head(v_norm, n = 10)## [1] -1.10087437 1.28091634 1.51635608 -2.67190898 2.76741063 1.37167187## [7] -3.43942011 -1.20752287 0.07888408 -0.813003819 / 25
take a minute to discuss with others
Coercion on purpose
as.integer(v_log2)## [1] 1 0 1 1 0as.character(v_dbl)## [1] "1" "3.1"as.logical(v_int)## [1] FALSE TRUE TRUE TRUEas.numeric(v_log2)## [1] 1 0 1 1 0as.numeric(v_chr)## Warning: NAs introduced by coercion## [1] NA NA10 / 25
Vector names
Three ways to add names
(v1 = c(a = 1, b = 2)) # 1## a b ## 1 2v2 = 1:2names(v2) = c("a", "b") # 2v2## a b ## 1 2setNames(1:2, c("a", "b")) # 3## a b ## 1 211 / 25
Vector names
Three ways to add names
(v1 = c(a = 1, b = 2)) # 1## a b ## 1 2v2 = 1:2names(v2) = c("a", "b") # 2v2## a b ## 1 2setNames(1:2, c("a", "b")) # 3## a b ## 1 2Remove names
unname(v1)## [1] 1 2names(v2) = NULLv2## [1] 1 211 / 25
Lists
Lists are different from atomic vectors above because their elements can be of any type, including lists (thus they are recursive vectors)
x = list(1:3, "a", c(TRUE, FALSE), list(2:1, "b"))str(x)## List of 4## $ : int [1:3] 1 2 3## $ : chr "a"## $ : logi [1:2] TRUE FALSE## $ :List of 2## ..$ : int [1:2] 2 1## ..$ : chr "b"is.recursive(x)## [1] TRUE12 / 25
Lists
l1 = list(list(1, 2), c(3, 4))str(l1)## List of 2## $ :List of 2## ..$ : num 1## ..$ : num 2## $ : num [1:2] 3 4l2 = c(list(1, 2), c(3, 4))str(l2)## List of 4## $ : num 1## $ : num 2## $ : num 3## $ : num 413 / 25
Lists
l1 = list(list(1, 2), c(3, 4))str(l1)## List of 2## $ :List of 2## ..$ : num 1## ..$ : num 2## $ : num [1:2] 3 4l2 = c(list(1, 2), c(3, 4))str(l2)## List of 4## $ : num 1## $ : num 2## $ : num 3## $ : num 4typeof(l1)## [1] "list"unlist(l1) # back to atomic vector## [1] 1 2 3 413 / 25
List names
names(l2)## NULLnames(l2) = c("name_1", "name_2")str(l2)## List of 4## $ name_1: num 1## $ name_2: num 2## $ NA : num 3## $ NA : num 4l3 = list(lst_a = c(1:5), lst_b = letters[1:3], LETTERS[1:3])str(l3)## List of 3## $ lst_a: int [1:5] 1 2 3 4 5## $ lst_b: chr [1:3] "a" "b" "c"## $ : chr [1:3] "A" "B" "C"names(l3)## [1] "lst_a" "lst_b" ""14 / 25
Matrix
matrix(data = 0, nrow = 3, ncol = 3)## [,1] [,2] [,3]## [1,] 0 0 0## [2,] 0 0 0## [3,] 0 0 0matrix(data = 1:9, nrow = 3, ncol = 3)## [,1] [,2] [,3]## [1,] 1 4 7## [2,] 2 5 8## [3,] 3 6 915 / 25
Matrix
matrix(data = 0, nrow = 3, ncol = 3)## [,1] [,2] [,3]## [1,] 0 0 0## [2,] 0 0 0## [3,] 0 0 0matrix(data = 1:9, nrow = 3, ncol = 3)## [,1] [,2] [,3]## [1,] 1 4 7## [2,] 2 5 8## [3,] 3 6 9matrix(data = letters[1:9], nrow = 3, ncol = 3)## [,1] [,2] [,3]## [1,] "a" "d" "g" ## [2,] "b" "e" "h" ## [3,] "c" "f" "i"matrix(data = LETTERS[1:9], nrow = 3, ncol = 3)## [,1] [,2] [,3]## [1,] "A" "D" "G" ## [2,] "B" "E" "H" ## [3,] "C" "F" "I"15 / 25
Matrix
mat_a <- matrix(data = 1:9, nrow = 3, ncol = 3, byrow = TRUE )mat_a## [,1] [,2] [,3]## [1,] 1 2 3## [2,] 4 5 6## [3,] 7 8 9rownames(mat_a) <- c("row1", "row2", "row3")colnames(mat_a) <- c("col1", "col2", "col3")mat_a## col1 col2 col3## row1 1 2 3## row2 4 5 6## row3 7 8 916 / 25
Matrix
Coercion
mat_b <- mat_amat_b[9] = "n9"mat_b## col1 col2 col3## row1 "1" "2" "3" ## row2 "4" "5" "6" ## row3 "7" "8" "n9"class(mat_b)## [1] "matrix" "array"typeof(mat_b)## [1] "character"17 / 25
matrix also has type conversion
Matrix
upper.tri(mat_a, diag = FALSE)## [,1] [,2] [,3]## [1,] FALSE TRUE TRUE## [2,] FALSE FALSE TRUE## [3,] FALSE FALSE FALSEmat_a## col1 col2 col3## row1 1 2 3## row2 4 5 6## row3 7 8 9(idx = lower.tri(mat_a, diag = TRUE))## [,1] [,2] [,3]## [1,] TRUE FALSE FALSE## [2,] TRUE TRUE FALSE## [3,] TRUE TRUE TRUEmat_a[idx]## [1] 1 4 7 5 8 918 / 25
Arrays
a = array(data = 1:12, dim = c(2, 3, 2))a## , , 1## ## [,1] [,2] [,3]## [1,] 1 3 5## [2,] 2 4 6## ## , , 2## ## [,1] [,2] [,3]## [1,] 7 9 11## [2,] 8 10 12length(a)## [1] 12dim(a)## [1] 2 3 2str(a)## int [1:2, 1:3, 1:2] 1 2 3 4 5 6 7 8 9 10 ...class(a)## [1] "array"typeof(a)## [1] "integer"19 / 25
Arrays: dimension names
dimnames(a) = list(c("R1", "R2"), c("C1", "C2", "C3"), c("A", "B"))a## , , A## ## C1 C2 C3## R1 1 3 5## R2 2 4 6## ## , , B## ## C1 C2 C3## R1 7 9 11## R2 8 10 1220 / 25
Arrays: dimension names
dimnames(a) = list(c("R1", "R2"), c("C1", "C2", "C3"), c("A", "B"))a## , , A## ## C1 C2 C3## R1 1 3 5## R2 2 4 6## ## , , B## ## C1 C2 C3## R1 7 9 11## R2 8 10 12a2 = array(data = 1:12, dim = c(2, 3, 2), dimnames = list(c("R1", "R2"), c("C1", "C2", "C3"), c("A", "B")))a2## , , A## ## C1 C2 C3## R1 1 3 5## R2 2 4 6## ## , , B## ## C1 C2 C3## R1 7 9 11## R2 8 10 1220 / 25
How the three objects below are different from vector 1:5?
x1 = array(1:5, c(1, 1, 5))x2 = array(1:5, c(1, 5, 1))x3 = array(1:5, c(5, 1, 1))21 / 25
Data frames
A data frame is more general than a matrix in that different columns can be different modes of data; it will be the most common data structure we'll deal with in R.
d = data.frame(v_dbl, v_log, v_chr)d## v_dbl v_log v_chr## 1 1.0 TRUE a## 2 3.1 FALSE wordstr(d)## 'data.frame': 2 obs. of 3 variables:## $ v_dbl: num 1 3.1## $ v_log: logi TRUE FALSE## $ v_chr: chr "a" "word"length(d)## [1] 322 / 25
Data frames
A data frame is just a list of equal-length vectors; therefore it shares properties of both matrix and list
d## v_dbl v_log v_chr## 1 1.0 TRUE a## 2 3.1 FALSE word# a list of equal length vectortypeof(d)## [1] "list"class(d)## [1] "data.frame"is.data.frame(d)## [1] TRUEnames(d)## [1] "v_dbl" "v_log" "v_chr"colnames(d)## [1] "v_dbl" "v_log" "v_chr"rownames(d)## [1] "1" "2"23 / 25
as.data.frame()
as.data.frame(c(1:2))## c(1:2)## 1 1## 2 2as.data.frame(mat_a)## col1 col2 col3## row1 1 2 3## row2 4 5 6## row3 7 8 9as.data.frame(l2)## name_1 name_2 NA. NA..1## 1 1 2 3 424 / 25
Combine data frames
stack data frames
d_row = data.frame(1, 2, "3")names(d_row) = names(d)rbind(d, d_row)## v_dbl v_log v_chr## 1 1.0 1 a## 2 3.1 0 word## 3 1.0 2 3dplyr::bind_rows(d, d_row)## v_dbl v_log v_chr## 1 1.0 1 a## 2 3.1 0 word## 3 1.0 2 3data frames side by side
d_col = data.frame(x1 = 1:2)cbind(d, d_col)## v_dbl v_log v_chr x1## 1 1.0 TRUE a 1## 2 3.1 FALSE word 2dplyr::bind_cols(d, d_col)## v_dbl v_log v_chr x1## 1 1.0 TRUE a 1## 2 3.1 FALSE word 225 / 25