Homework assignments
At this moment, you should have installed all required tools on your computer. You also should have some general ideas about what they can do. In this assignment, you will:
- Register a GitHub account and create a new private repository named as
BIOL7800. - Within the repository, add an RStudio project file; you can name it as
biol7800.Rproj. - Create a folder named as
homework; and then a subfolder named ashw_01. - Within
hw_01, create a RMarkdown file named ashw_01.Rmd. You can use the default template for it. - Replace the content with a short self introduction of yourself. Try to use some markdown syntax (e.g., section title, bold, italic fonts, etc.).
- Also insert bash chunks to print out the versions of
gitandRinstalled on your computer.
After done, compile the Rmd file to html and pdf. Push all files (Rmd, html, and pdf) to your BIOL7800 repository on GitHub. You need to add me as a collaborator for your repository. This repository is where you are going to submit all homework assignments and final reports. I will grade it by cloning it to my computer and recompile it. The last commit time will be used to check whether it is late or not. Remember, no late homework will be accepted. If the last commit time is after the deadline, I will use whatever commit that was before the deadline as your submission.
This homework will be due at September 14th, 9am.
- (2 points) Fix each of the following common data frame subsetting errors:
mtcars[mtcars$cyl < 6] mtcars[-1:3,] mtcars[mtcars$cyl = 8, ] mtcars[mtcars$cyl == 4 | 6, ] - (1 point) Why does the following code generated five missing values?
x = 1:5 x[NA] - (2 points) Why does
mtcars[1:15]return an error? How does it differ frommtcars[1:15, ]? - (2 points) Explain how does the following code work.
x = matrix(c(1:3, NA, 5:7, NA, NA), nrow = 3) x[is.na(x)] = 0 - (3 points) Load the Car Road Tests dataset (in R, run
data("mtcars"),?mtcars), then add a new column named asmpg_2for themtcarsdata frame. You can useif ... else ...orifelseor any other functions that can get the job done. This new column will categorizempginto four categories using the thresholds below:mpg_2categoryThresholds Low mpg < 16 Low_intermediate 16 <= mpg < 21 Intermediate_high 21 <= mpg < 26 High 26 <= mpg
This homework will be due at September 28th, 9am. Please submit your homework as an Rmd file to GitHub. It is probably a good idea to create a hw_02 subfolder within your homework folder and put the Rmd file there. Please also generate a PDF file there. You don’t need to email me again about your homework. I have your GitHub repo link and will check out your homework there. This should apply to all homework assignments.
-
(2 points) Suppose we have a dataset A (see code below) where each column represents multiple measures of nitrogen concentration in a particular lake. We want to get the average value for each lake. Do this in two ways: a
forloop and a vectorized functioncolMeans().set.seed(12) # to be reproducible A = matrix(data = runif(n = 1:500), nrow = 50, ncol = 10) colnames(A) = paste("lake", 1:10, sep = "_") -
(2 points) From the for loop lecture, we see the following example of using
apply():x = array(1:27, dim = c(3, 3, 3)) apply(X = x, MARGIN = c(1, 2), FUN = paste, collapse = ", ")Now, use
forloops to get the same task done (hint: nested loops). The results should be the same. -
(2 points) The Fibonacci Sequence is the series of numbers that the next number is the sum of the previous two numbers: 0, 1, 1, 2, 3, 5, 8 … Use a
forloop to get the first 30 numbers of the Fibonacci Sequence. This question should demonstrate the need for loops because there is no easy way to use vectorized functions in this case. -
(2 points) In the example data below, extract those ranking numbers with regular expression. The results should have the number(s) and
.if it follows after the numbers immediately (i.e.,1.,12.,105.,105.3, etc.). Remove empty strings from the final results. You should get 107 strings for your results.top105 = readLines("http://www.textfiles.com/music/ktop100.txt") top105 = top105[-c(64, 65)] # missing No. 54 and 55 -
(2 points) For the vector with length of 107 you got from question 4, remove all trailing
.. (hint:?sub). Then convert it to a numeric vector and find out which numbers have duplications (i.e., a tie in ranking). Don’t count by eyes, use R to find it out (hint:table(),sort(); orduplicated(),which(),[subsetting; there are more than one way to do so).
This homework will be due at October 12th, 9am.
- (3 points) Use the
rvestR package to scrape the schedule and materials table into R from the course webpage (https://introdatasci.dlilab.com/schedule_materials/). Read the documentation ofrvestso you get a better idea about the functions provided byrvestand their usages. - (2 points) With the extracted data frame, create two new columns based on the
Datecolumn:monthandday.monthwould be the month abbrevations from theDatecolumn;daywould be the numeric numbers from theDatecolumn. Although you can use whatever approach to get this done (do not enter them by hand…), I suggest you try to practice regular expression here (sub()orstringr::str_extract()). - (2 points) With the data frame generated from Q2, use
group_by()andsummarise()to find out the number of lectures for eachmonth, order the results by the number of lectures (high to low). - (3 points) For the
Topiccolumn, split all values into words (hint:stringr::str_split()). Observe the values in theTopiccolumn and use regular expression to specify thepatternin thestringr::str_split()orstrsplit()function. Once this is done, you should get a list of list, you can useunlist()to convert it into a vector and name it aswords. Usetable()andsort()to find the top 5 most frequent words.
I was thinking to have a homework to get all plant occurrence data within Baton Rouge from GBIF. But it will require you to register an account and have account name and password when you use the rgbif package. This may have the risk of get your password leaked (you can avoid this by reading the documentation of rgbif); so I decided not to do so. If you are interested, you may want to run some example codes from the rgbif package’s documentation.
This homework will be due at October 31th, 9am.
In the neonDivData data package, there is a data frame named as data_plant. This data frame records plant coverage (percentage at 1 m^2^ scale indicated by the sample_area_m2 column) and plant presence information in larger plots (10 and 100 m^2^ indicated by the sample_area_m2 column). Use this data frame and functions we learned during lectures to do the steps below.
-
(2 points) Create a new column named as
genusfordata_plantfrom thetaxon_namecolumn. The genus name is the first word of the scientific names. For example, if a record hastaxon_nameof"Bunchosia glandulosa (Cav.) DC.", then the genus is"Bunchosia". You probably want to use regular expression to do so. Take a look at all the names (sort(unique(data_plant$taxon_name))) to look at possible genus names and think about how to specify the regular expression pattern. Randomly select 100 values from thegenuscolumn and print it out. -
(2 points) Looking at the
taxon_namevalues, it is clear that some scientific names probably are the same species (as different subspecies). For example, we may want to treat “Calamagrostis canadensis (Michx.) P. Beauv.” and “Calamagrostis canadensis (Michx.) P. Beauv. var. langsdorffii (Link) Inman” as the same species. Create a new columntaxon_name2fordata_plantbased ontaxon_name.taxon_name2should just contain the first two words oftaxon_name. For example, “Calamagrostis canadensis (Michx.) P. Beauv.” and “Calamagrostis canadensis (Michx.) P. Beauv. var. langsdorffii (Link) Inman” should both be “Calamagrostis canadensis”. Randomly select 100 values from thetaxon_name2column and print it out. -
(2 points) Calculate the number of species (based on
taxon_name2) of each site observed based on different sizes of plot:- based on 1 m^2^ plots; this would be all observations with
sample_area_m2 == "1". This would result in a data frame named asn_1with two columns:siteIDandrichness_1m2. - based on 10 m^2^ plots; this would be all observations with
sample_area_m2 %in% c("1", "10"). This would result in a data frame named asn_10with two columns:siteIDandrichness_10m2. - based on 100 m^2^ plots; this would be all observations with
sample_area_m2 %in% c("1", "10", "100"). This would result in a data frame named asn_100with two columns:siteIDandrichness_100m2.
then, use
dplyr::left_join()to joinn_1,n_10, andn_100as one data framen_all, which should have 47 rows and four columns:siteID,richness_1m2,richness_10m2, andrichness_100m2. Note:dplyr::left_join()can only join two data frames at each time, so you may use pipe (e.g.,xyz = left_join(x, y) %>% left_join(z)). - based on 1 m^2^ plots; this would be all observations with
-
(2 points) Transform
n_allto a long format data frame named asn_all_longwith three columns:siteID,spatial_scale, andrichness. Hint:tidyr::pivot_longer(). -
(2 points) Use
ggplot2andn_all_longto generate the plot below. Each line links the three values of each site (hint:aes(group = siteID)).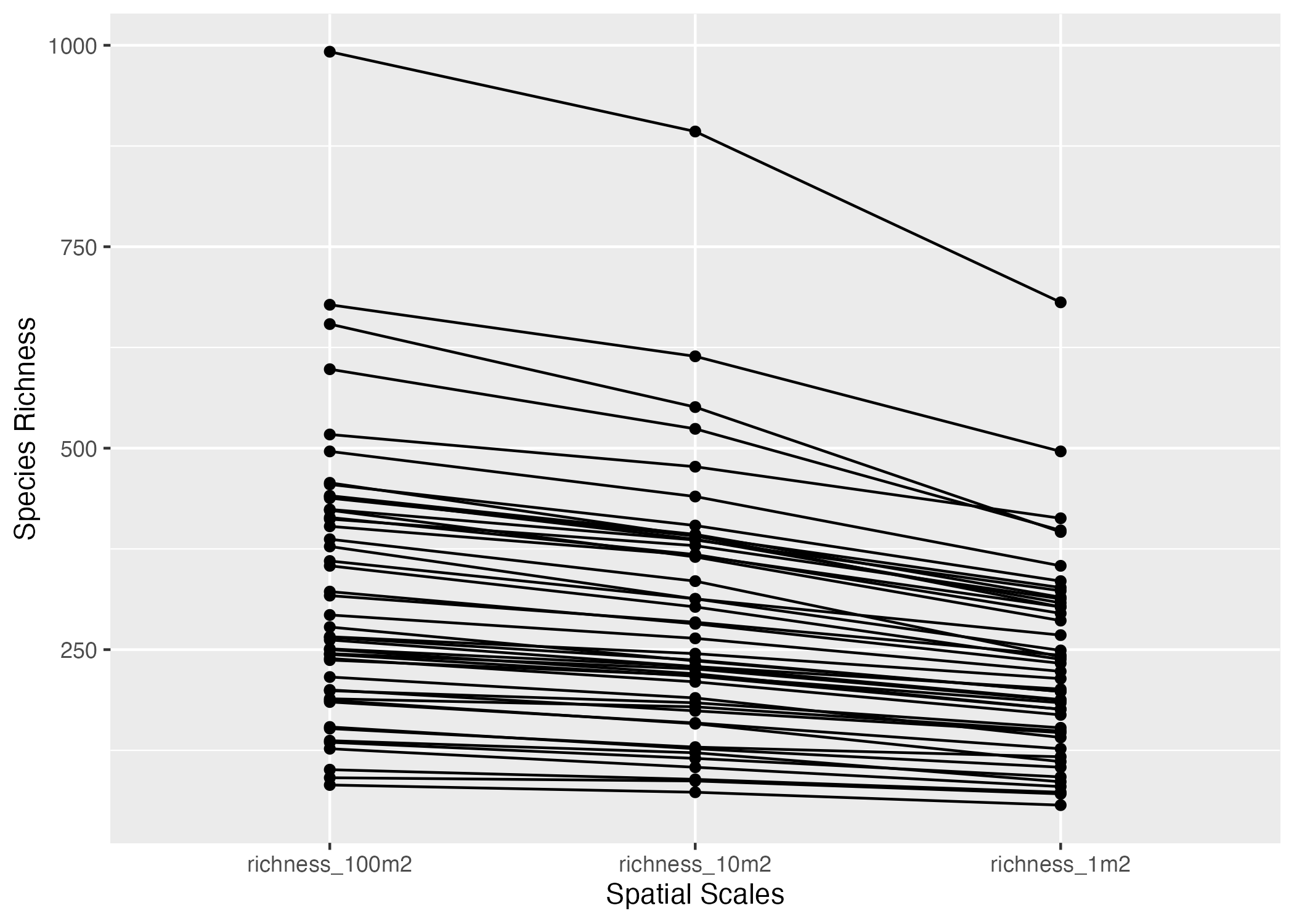
This homework will be due at November 2nd, 9am.
Note: Please please please write some text in your answers, do not just put R code there! Also, you should have a PDF file along with the Rmd file in your homework folder. For this homework, you can use R to get answers except 1c, where you should have a hand-calculated solution (show the process); you can use R code to verify your answer though. If you assigned anything as an object in R for your answers, remember to print it out so that I can see your results.
The data consist of a predictor variable x, plant height, and a response variable y, grain yield, for eight varieties of rice.
x = c(110.5, 105.4, 118.1, 104.5, 93.6, 84.1, 77.8, 75.6)
y = c(5.755, 5.939, 6.010, 6.545, 6.730, 6.750, 6.899, 7.862)
Consider fitting a simple linear regression model where , i = 1, 2, …, 8.
- (1 point) Give the least squares estimate () of the slope . Give a brief interpretation of .
- (1 point) Perform a test for versus using an F test first and then a T test. Your conclusion?
- (1 point) Construct a 95% confidence interval for the intercept by hand using the equation from the lecture, compare your results with those from R and briefly interpret the 95% confidence interval. You can get using R code
qt(alpha/2, n-2)where alpha is 0.05 here. - (1 point) Give the fitted regression line (as a equation that looks like ) and the raw residuals.
- (1 point) Give an estimate () of the error variance ().
- (1 point) Estimate the expected yield of a rice variety () that has height and provide a 95% confidence interval.
- (1 point) Predict the yield of a new rice variety that has height and provide a 95% prediction interval. Compare the results with those from (f), which one is wider?
- (1 point) Compute the coefficient of determination and briefly interpret what does it mean.
-
This problem is designed to demonstrate why residuals are plotted against (instead of ). Consider the following (artificial) data set that was constructed so that the relationship between and is quadratic. It is immediately evident that a linear fit is not appropriate. However, we adopt the point of view that the residual plot will provide diagnostic information on the lack of fit.
x = c(1, 2, 3, 4, 5, 6, 7, 8, 9) y = c(-2.08, -0.72, 0.28, 0.92, 1.20, 1.12, 0.68, -0.12, -1.28)
(0.25 point) Plot vs. .
(0.25 point) Plot the raw residuals vs. .
(0.25 point) Plot the raw residuals vs. .
(0.25 point) Plot the raw residuals vs. .
(1 point) Compare the plots from (b), (c), and (d). Is there a meaningful difference between (c) and (d)? Explain. Which of the plots (b) or (d) gives a better indication of the lack of fit? Explain.
Note: If you were to compute the correlation between and the raw residuals, you would find this to be 0. If you compute the correlation between the observed and the raw residuals, you find that this is where . It is the absence of correlation in plot (d) — and also plot (c) — that is important. No computations of correlation are required in this problem, but you may give it a try in R.
This homework will be due at November 16th, 9am.
-
(2 points) Reorganize your
BIOL7800Github Repository to have the following structure. If your respository already has this structure, great!! You will get all the points for this question.. ├── final_project └── homework ├── hw_01 ├── hw_02 ├── hw_03 ├── hw_04 ├── hw_05 ├── hw_06 └── hw_07 -
(8 points) Create an R package and push it to your Github account. You can name it whatever you want. In the package, put two functions that you have written or you want to write. If you don’t have such functions or ideas, try this: one function to remove rows with all 0s in the matrix generated below; and the other function to remove columns with all 0s. Name the first one as
rm_0s_by_row()and the other asrm_0s_by_col().set.seed(123) x = matrix(rpois(100, 0.1), 10, 10)Document them! I should be able to install your package using
remotes::install_github("your_github_name/pkg_name")and run your code and read documentations.You can work out the package locally first. Once success (pass
devtools::check(); no error, no warning, no note), go to Github, create a new repository, don’t add anything there yet (i.e., no README or license file added by Github). Then follow the command it provides and link your local directory to the Github repository; then commit and push it to Github repository.Put the link to your R package Github repository in the
hw_07.mdfile that will be found in yourBIOL7800/homework/hw_07folder (so that I know where it is).
This homework will be due at November 30th, 9am.